Introduction
Cardiac fibroblasts are key players in remodeling after myocardial infarction (MI) and could be a therapeutic target for treatment (Chen & Frangogiannis, 2013). They are known to secrete various extracellular matrix proteins that are involved in a stable scar formation. Previous studies of our group using stable isotopes (SILAC) have shown that MI-activated fibroblasts (aCF) in culture can also secrete numerous paracrine factors like cytokines, chemokines, growth factors suggesting that they may influence inflammation and may be cardioprotective. However, culture conditions could have critically influenced the results obtained and therefore may not be directly extrapolated to in vivo conditions. In the present study, we report the first secretome analysis of aCF under in vivo conditions using a novel proximity ligation technique to specifically label secreted proteins in aCF.
Method
Cardiac fibroblasts are key players in remodeling after myocardial infarction (MI) and could be a therapeutic target for treatment (Chen & Frangogiannis, 2013). They are known to secrete various extracellular matrix proteins that are involved in a stable scar formation. Previous studies of our group using stable isotopes (SILAC) have shown that MI-activated fibroblasts (aCF) in culture can also secrete numerous paracrine factors like cytokines, chemokines, growth factors suggesting that they may influence inflammation and may be cardioprotective. However, culture conditions could have critically influenced the results obtained and therefore may not be directly extrapolated to in vivo conditions. In the present study, we report the first secretome analysis of aCF under in vivo conditions using a novel proximity ligation technique to specifically label secreted proteins in aCF.
Method
For proximity labeling we used TurboID which is a biotin ligase derived from E. coli (Branon et al., 2018) and catalyzes the binding of biotin to proximate proteins. This allows the analysis of biotinylated proteins of a specific cell type and has been recently described for hepatocytes (Wei et al., 2021). Here we adapted this method for aCF. As shown in figure 1, an adeno associated virus of serotype 9 (AAV9) carrying a plasmid with the TurboID sequence under control of a Periostin (Postn) promoter was injected into the tail vein of C57Bl/6 mice to specifically target aCF within the heart. Myocardial infarction (50 min ischemia/reperfusion) was induced and TurboID expressed in aCF as a result. For TurboID mediated biotin labeling of proximate proteins, biotin was applied to the mice via drinking water. Coronary effluent perfusate of Langendorff perfused hearts was collected and biotinylated proteins enriched using streptavidin beads to analyze secreted proteins by mass spectrometry.
Workflow
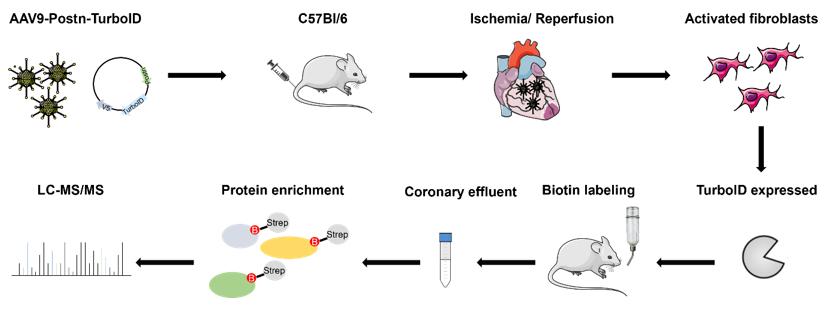
Figure 1: Workflow of in vivo secretome analysis for activated cardiac fibroblasts (aCF) according to Wei et al. 2021.
Workflow
Figure 1: Workflow of in vivo secretome analysis for activated cardiac fibroblasts (aCF) according to Wei et al. 2021.
Results
Control experiments using immune fluorescence imaging revealed that TurboID was exclusively expressed in activated cardiac fibroblasts after MI. Analysis of the biotinylated proteins 5d post MI identified several extracellular matrix proteins including Fibronectin (Fn1), Heparan sulfate proteoglycan 2 (Hspg2), Nidogen-1 (Nid1), Laminin subunit gamma 1 (Lamc1), Laminin subunit alpha 4 (Lama4) and Laminin subunit alpha 2 (Lama 2). Interestingly, we also identified paracrine factors such as Clusterin (Clu) Thrombospondin 4 (Thbs4), Fibulin-2 (Fbln2) and Gelsolin (Gsn) which have been previously reported to be cardioprotective (van Dijk et al., 2010, Doroudgar & Glembotski, 2013). The specificity
of the labeling process was further documented when genes coding for the
identified proteins were searched in the scRNAseq database recently reported by us (Hesse, Owenier et al., 2021): secreted proteins were exclusively
expressed in cardiac stromal cells (aCF and epicardial cells); furthermore, the
identified fibroblast clusters differentially contribute to the secretome.
Conclusion
We have successfully established a method which permits the in vivo secretome analysis of aCF in the infarcted heart. Aside of structural proteins, we have identified several secreted paracrine factors which are likely important in the protection of the injured myocardium. Thus, activated cardiac fibroblasts seem to be important players in cardioprotection.https://dgk.org/kongress_programme/jt2023/aP2203.html